An Idea About Embryonic Development
By: Nicole Toppses, Biochemistry, Class of 2023
Nominating Faculty Member: Professor Robert Morris
Our cells talk to each other as we grow from egg to person to coordinate our intricate design instructions. While we know the heavy-hitter in our design is DNA, the unsung heroes of the developmental process are cilia. What are cilia?
Cilia are cellular appendages extending from most of our cells. These complex structures serve both motor and sensory functions. For example, the respiratory system uses motile cilia to move mucus which traps dust and germs. Sensory cilia, or primary cilia, direct developmental signaling pathways, especially during embryonic development. These signals are molecules with interesting names, such as Notch, Wnt-Frizzled, and Sonic hedgehog (yes, this is the actual name). The unusual nature of these names does not take away from their importance. For example, the comically named sonic hedgehog signaling pathway controls the development of organs and the organization of our organ systems.
When cilia function abnormally or fail to function altogether, this causes diseases known as ciliopathies. Ciliopathies include polycystic kidney disease, which is the most common heritable genetic disease in humans. Ciliopathies also lead to brain defects, obesity, and bone abnormalities.
This project explores genetic patterns that drive cilia development in animals during embryonic development. The model system we study is the purple sea urchin. These fascinating creatures develop cilia within 24 hours of fertilization, making observation of cilia development easier. Surprisingly, their embryonic development is quite similar to the development of humans. Why is research on genetic patterns of cilia formation important? In order to treat ciliopathies, the disease-causing target must be accessible and identifiable. Mutations in the genes that encode the building blocks for cilia are the primary cause of ciliopathies. A deeper understanding of the genetic patterns controlling cilia formation in the purple sea urchin can bring new ideas to ciliopathy research and treatment.
Exploration of Cilia in Development
This project reviews and combines the work of Wheaton students that occurred from 2009 to 2019 to tell a story of cilia gene expression patterns. These genes include the building blocks, on-switches, and machines that build cilia during cilia formation, scientifically called ciliogenesis. When a gene turns on, we say it is “expressed.” Some genes encode proteins, called transcription factors, that are the on-switches for building block and machine gene expression. The pattern of turning on and off genes is the subject of this study.
The National Institute of Dental and Craniofacial Research (NIDCR) data on gene expression patterns over time represents the average of five expression level measurements for every sea urchin gene across five stages of development. These stages are the zygote (2 hr), early blastula (15 hr), late blastula (30 hr), gastrula (48 hr), and pluteus (72 hr). Embryonic development begins with the zygote, the fertilized egg. The zygote divides and transforms into a blastula which is a hollow ball of cells. Next, the hollow ball becomes a cup-like structure with three layers of cells known as endoderm, ectoderm, and mesoderm. These three groups of cells multiply and develop into different tissue types, which form our organs and organ systems. In sea urchins, the gastrula stage develops into the pluteus or larval stage. The sea urchin larva continues to develop and grow into the spiky purple ball we recognize as the adult purple sea urchin.
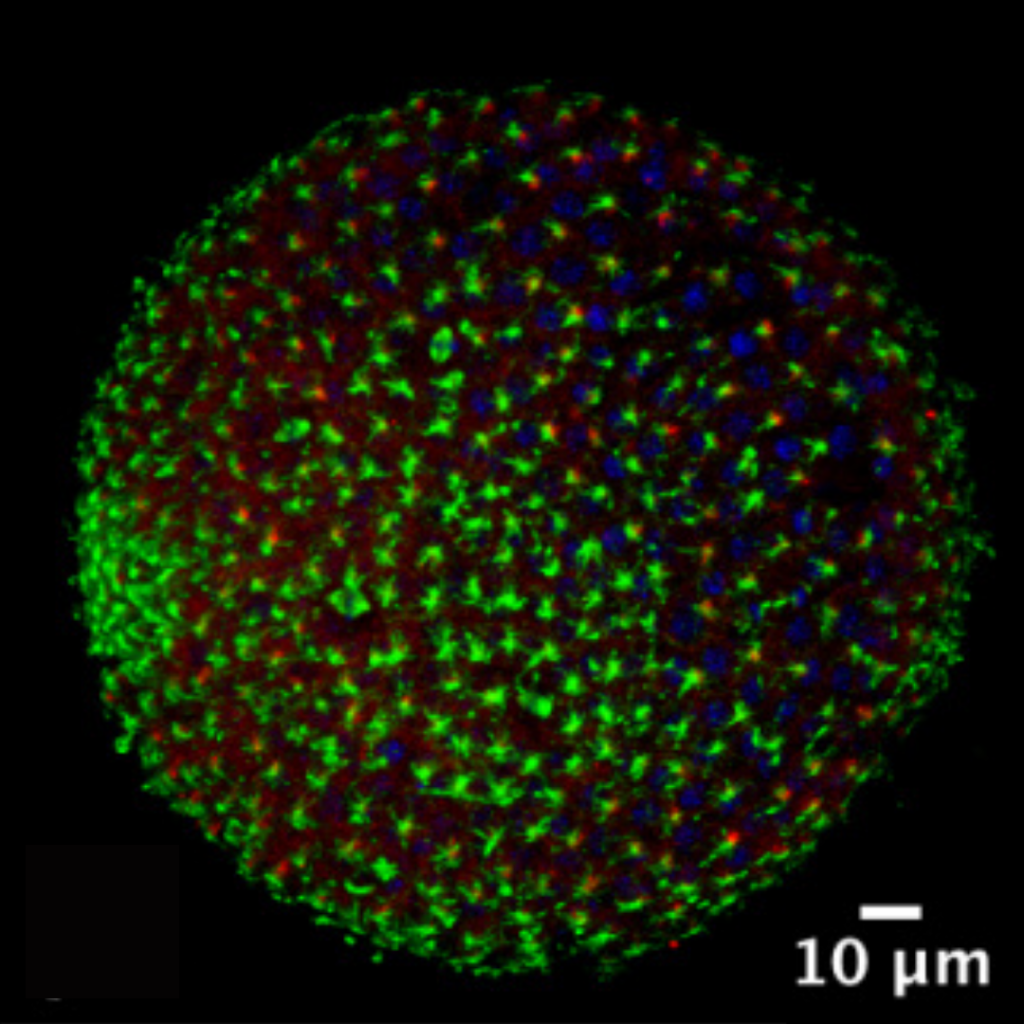
The predicted sea urchin ciliary gene data set consists of 317 unique potential cilia genes. These genes include lists of ciliary kinesin, tubulin, and dynein genes. The kinesin and dynein genes encode the protein machines that deliver the tubulin building blocks and make cilia. In the microscope image shown here (Figure 1), the building blocks proteins tubulin and acetylated tubulin show the cilia structure on the sea urchin during the late blastula stage.
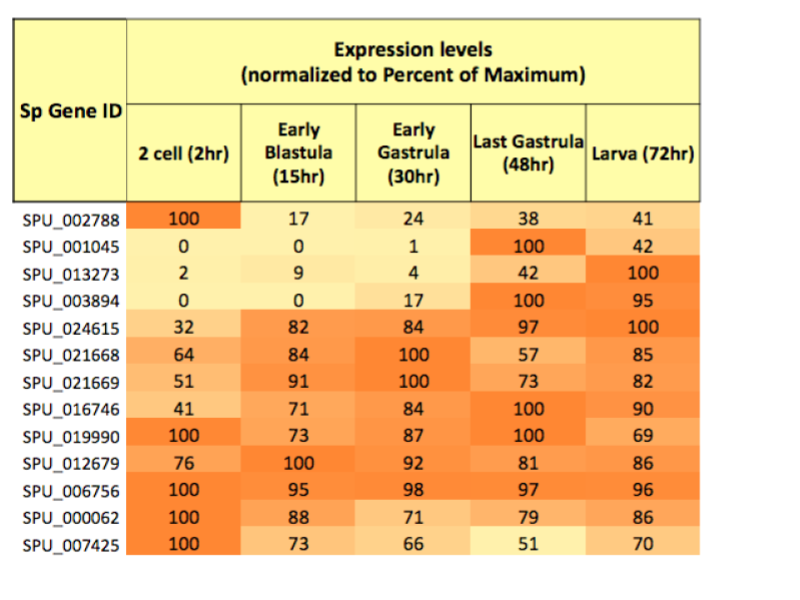
The patterns of change in gene expression were analyzed with a few different computer algorithms. The CLuster Identification via Connectivity Kernels (CLICK) computer algorithm identified categories of ciliary gene expression patterns over time. The CLICK algorithm suggested significant developmental expression levels in 191 of the 317 ciliary genes. Figure 2 shows the tubulin gene expression level patterns across the five identified stages of development. Since gene expression changes are controlled by transcription factors, we looked for transcription factor binding sites, or motifs, on the DNA. Motifs are repeating patterns in a genetic sequence where the transcription factors bind to and turn on genes. Analysis to identify potential motifs for ciliary genes expressed during the blastula stage used the creatively named computer programs MEME (Multiple EM for Motif Elicitation) and TOMTOM (a motif comparison tool in the MEME suite).
What This Means
The CLICK computer analysis shows that 168 of the 191 genes form four distinct expression groups or clusters, grouped by their highest expression level during the five developmental stages. The remaining 23 genes did not fit into the clusters and are called singletons. The expression clusters identified are during the zygote stage (2 hours), zygote-blastula (2-15 hours), early blastula (15 hours), and larval stages (72 hours). There is no cluster during the gastrula stage, suggesting that the cilia do not transform into different cilia types during this stage. The zygote cluster includes 62 genes, the zygote/blastula cluster represents 28 genes, the early blastula cluster represents 42 genes, and the larval cluster represents 37 genes. The genes of the blastula cluster may trigger the initial ciliogenesis event that occurs during the blastula stage. The types of genes identified in the blastula cluster include tools needed for initial cilia growth, supporting this hypothesis.
The MEME and TOMTOM analysis identified motifs in the blastula cluster that may be binding regions for transcription factors. These transcription factors turn on genes for ciliogenesis and cilia maturation events. The output from the TOMTOM algorithm included a motif for the binding site of the Glis3 transcription factor. Glis3 is a crucial transcription factor in turning on the hedgehog signaling pathway, suggesting its presence in the blastula cluster may direct the initial ciliogenesis event. The larval cluster genes are peculiar because their expression increases during cilia-differentiation events. In other words, this suggests they are essential in creating the different motile and primary cilia types during this late developmental stage. Knowing which cilia genes work together during development opens possibilities for future discoveries in ciliopathy research.
Acknowledgments
I would like to thank Wheaton Alumni Blair J. Rossetti (2009), Robert T. Manguso (2010), Madeline L. Keyes (2010), Javon R. Mullings (2016), and Kira E. Olander (2019), whose work set the foundation for this project and future research in the Morris Lab under the guidance and encouragement of Professor Robert Morris.
-
Categories:
- Academic Festival
- Bioinformatics
- Biology
- Genomics